How it works
Intro
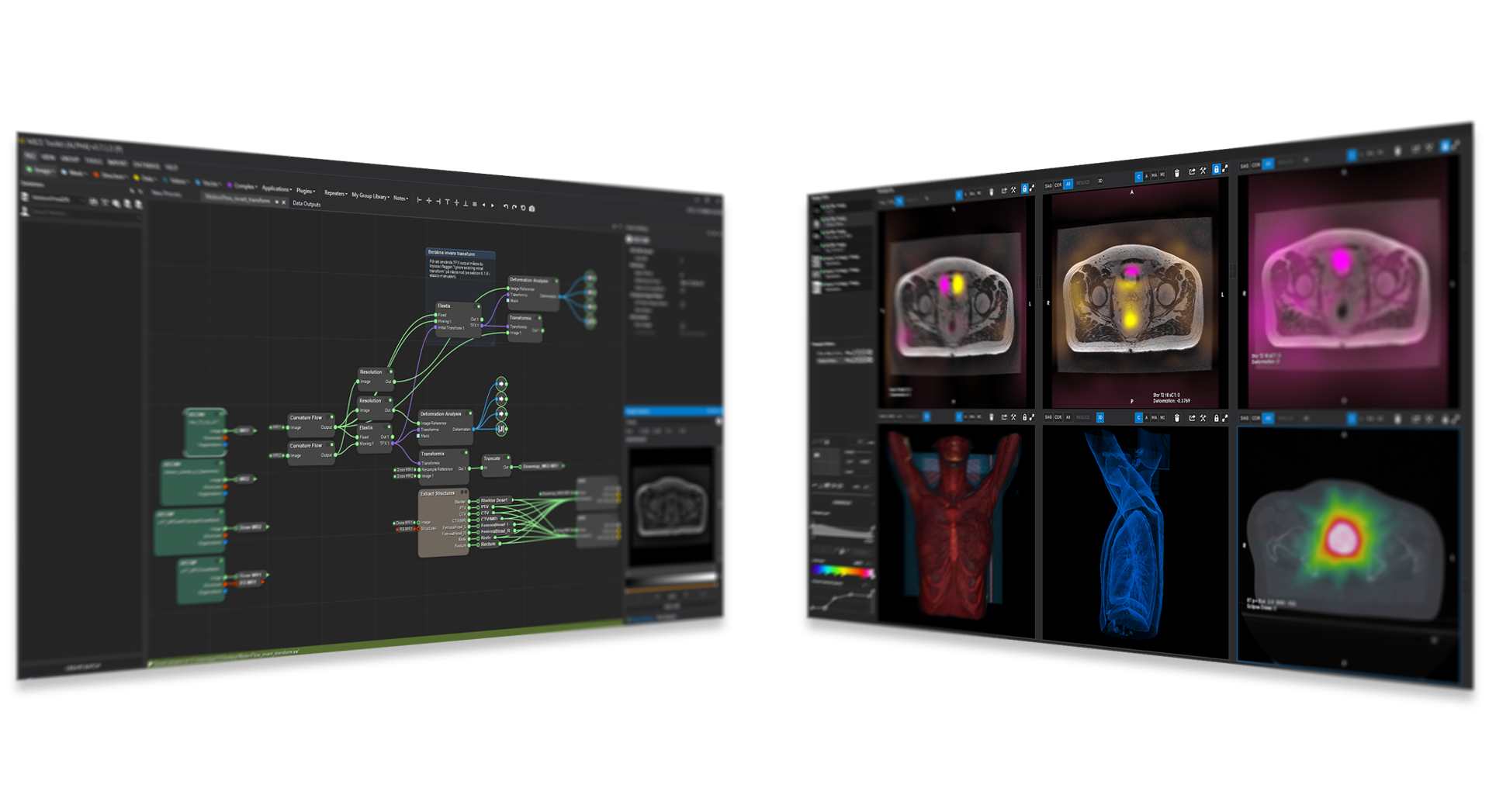
Image data
MICE Toolkit™ supports import and export of images in many file formats, including DICOM, NIfTI and MHD, as well as conventional image formats such as png, animated GIF files and STL files for 3D printing applications. DICOM and NIfTI files can be imported and structured in one or several databases with flexible tools for database handling. MICE Toolkit™ also supports importing and exporting DICOM RT Structures and importing DICOM RT Dose for radiotherapy
Building a workflow
Creating custom nodes
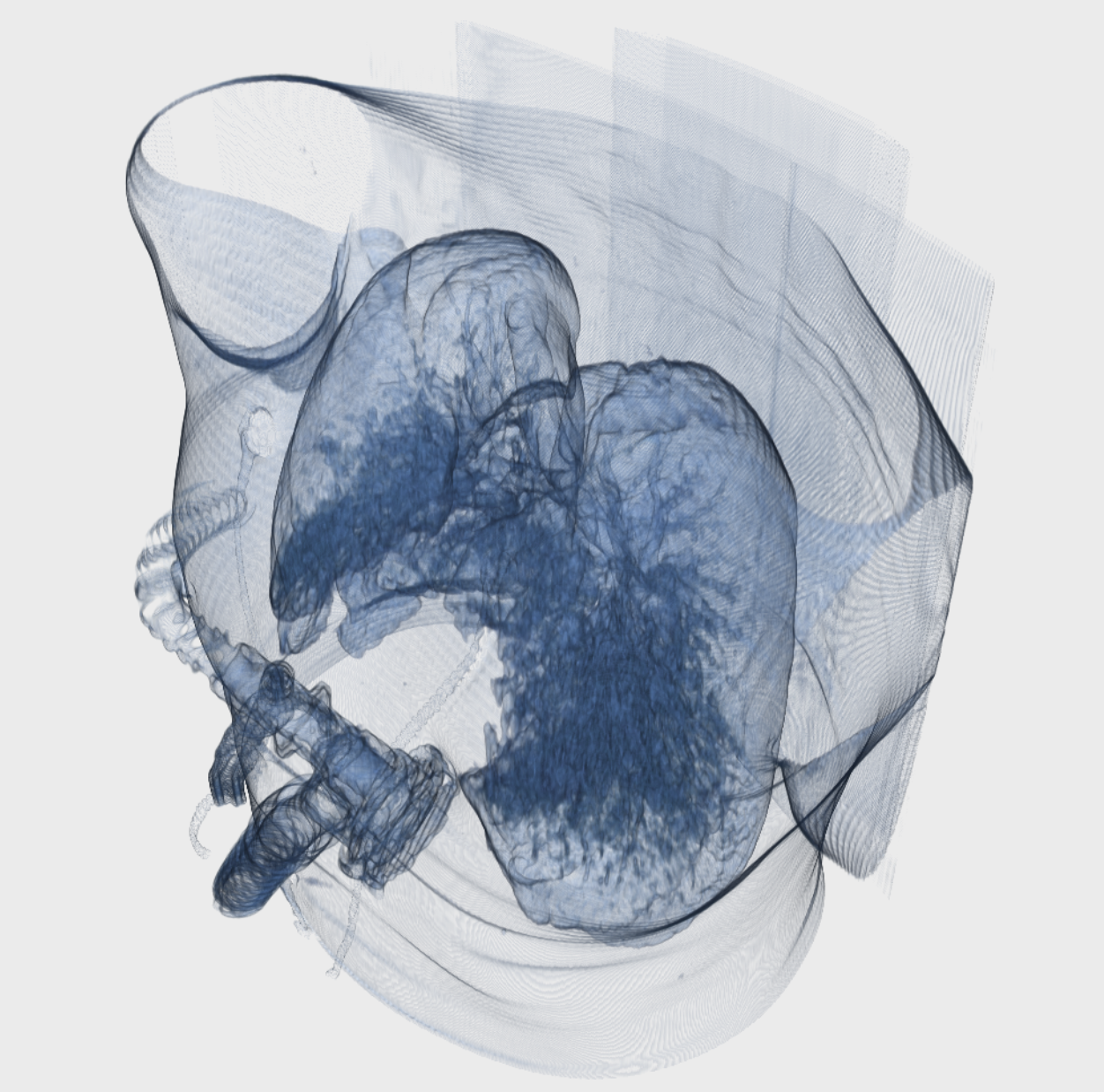
Visualization
The image visualizer in MICE Toolkit™ provides state-of-the-art visualization of medical image data, with up to six simultaneous viewports. Each viewport can show either 2D image projections or a 3D surface rendering, and viewport coordinate systems can be synchronized between viewports. The visualizer also provides tools for defining regions of interest that can be used in the pipeline or saved to a database and used at a later point in the analysis process.
Analysis in batches
Many analysis workflows are intended for application on a larger cohort of patients or images. To facilitate easy processing of large amount of data we provide batch analysis of workflows in MICE Toolkit™. This means that the workflow is applied to all images or patients that fulfil predefined criteria. The resulting images from a batch analysis can be visualized, saved to disk or to any database within MICE Toolkit™; and numerical values can be exported to .csv or .xlsx.
.files for each instance and for the entire cohort, for further statistical analysis. Each workflow in the batch is saved to disk, which makes it easy to retrospectively run workflows for any patient or image individually to verify the results or for debugging.
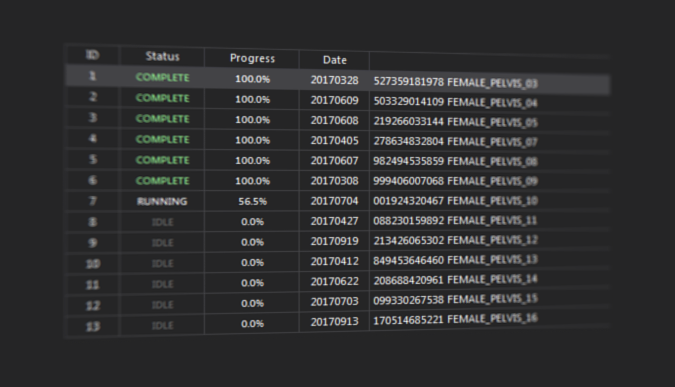
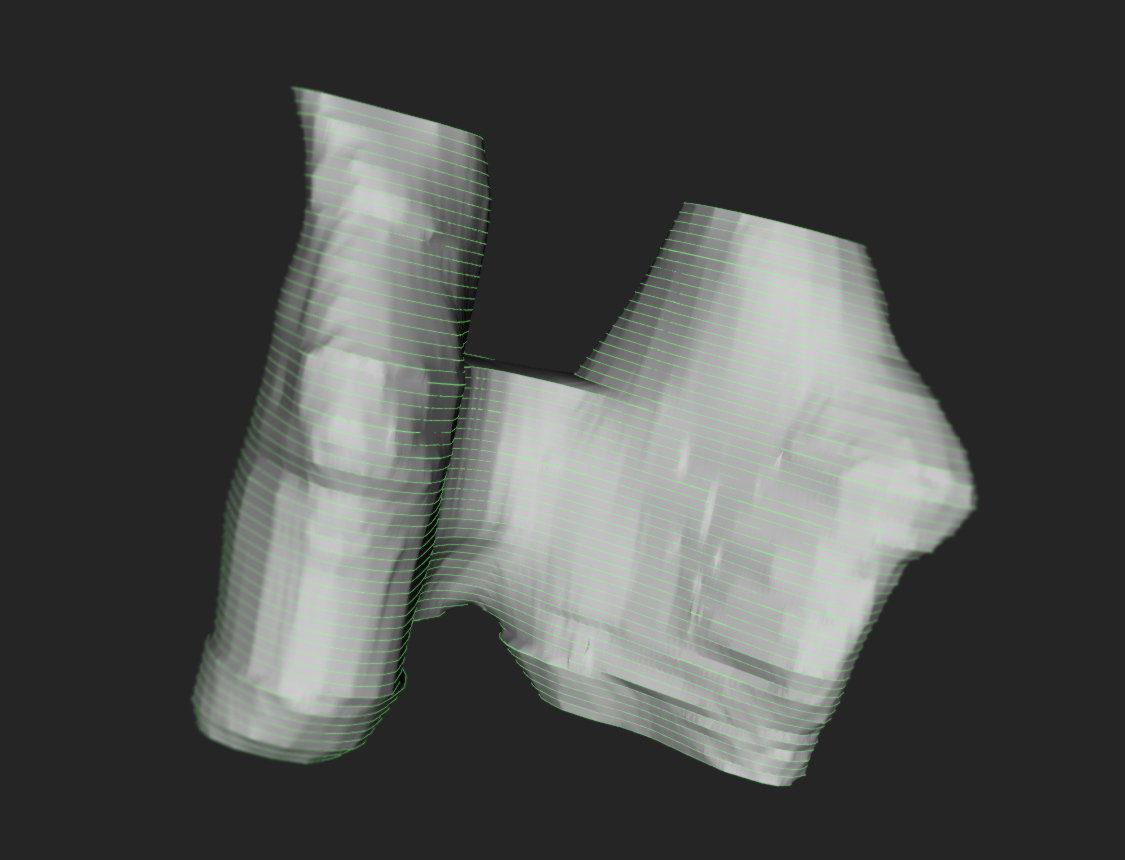
Extract results
